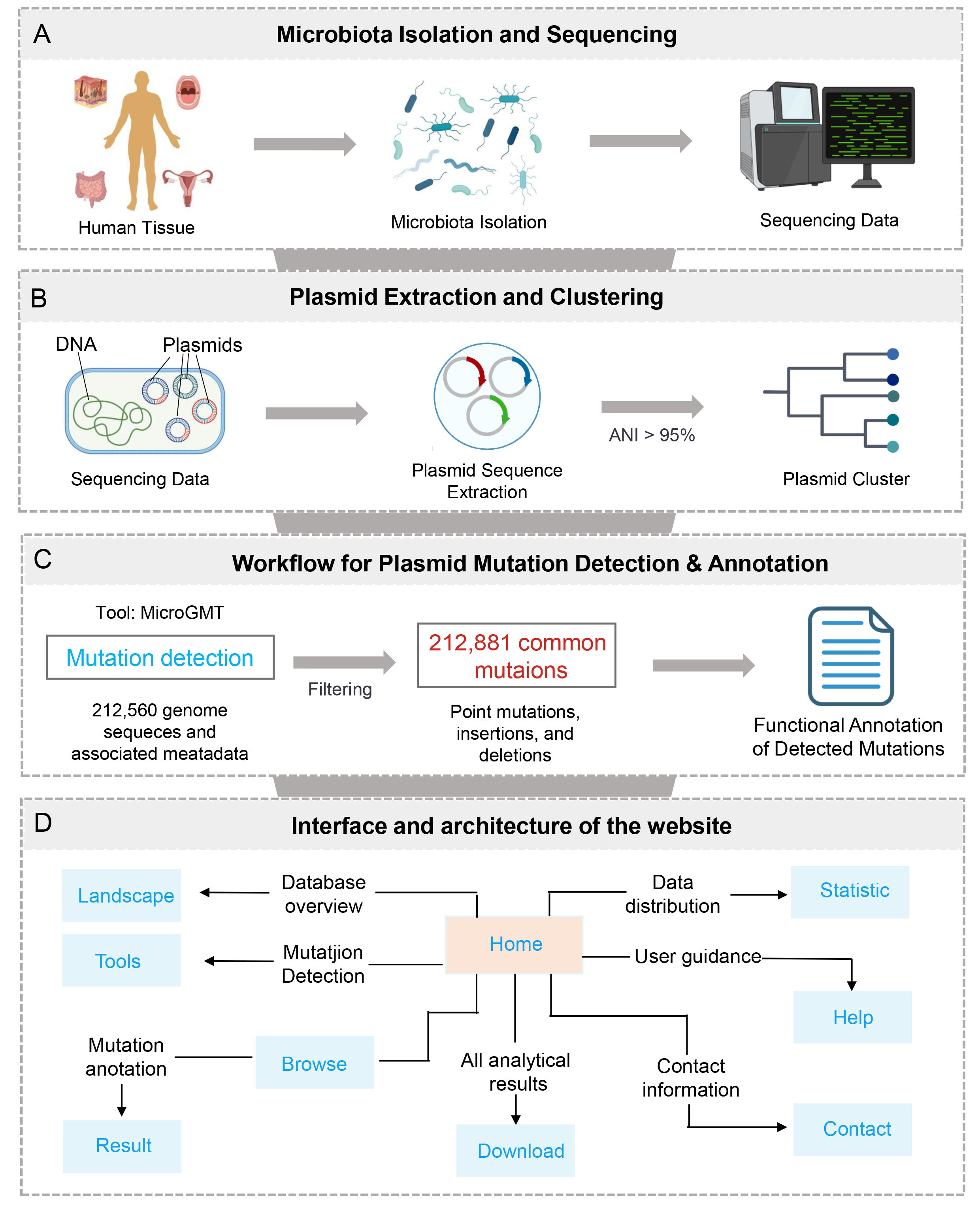
Plasmid Pipeline Overview.
Note: (A) Samples were collected from four body sites: skin, oral cavity, gut, and vagina. Microbial communities were obtained from each of these four sites. Metagenomic sequencing was performed on the collected samples. (B) Plasmid sequences were extracted from the metagenomic data using tools such as genomad, Platon, and Plasmer. Plasmid sequences were clustered using an average nucleotide identity (ANI) cutoff of > 95 %. (E) Mutations within the plasmid sequences were identified. (C) Plasmid mutation detection and annotation. Functional annotation of the mutations was conducted to assess their potential impact on plasmid function. (D) Interface and architecture of the website.